THINKCYTE SCIENCE
Label-free, high throughput cell characterization and isolation based on high-resolution morphology and artificial intelligence.
Ghost Cytometry, the Science-published technology inside VisionSort, brings artificial intelligence to cytometry to enable high-throughput, data rich, and novel insights for life science R&D.
With Ghost Cytometry, harness the power of AI in your cellular analysis and sorting workflows using both supervised and unsupervised machine learning approaches. Match the approach with your downstream R&D goals and experience ultimate control with powerful algorithms and visualization tools embedded directly in VisionSort.

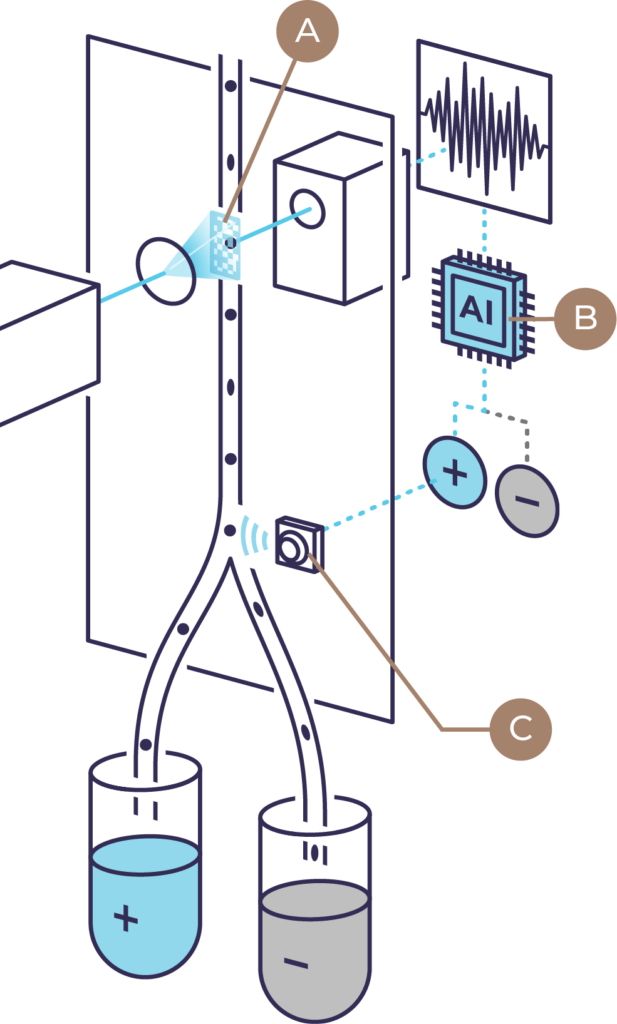
Morphological profiles are paired with fluorescently labeled markers for cell classes of interest.
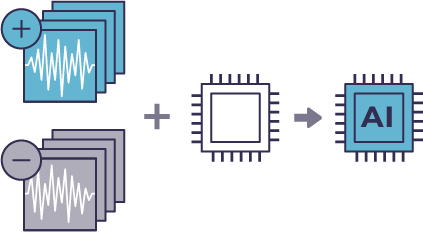
A machine-learning model is developed.
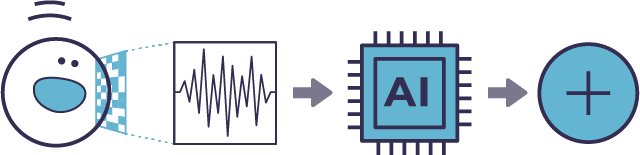
The machine-learning model predicts cell classes by evaluating morphological profiles in unlabeled samples.
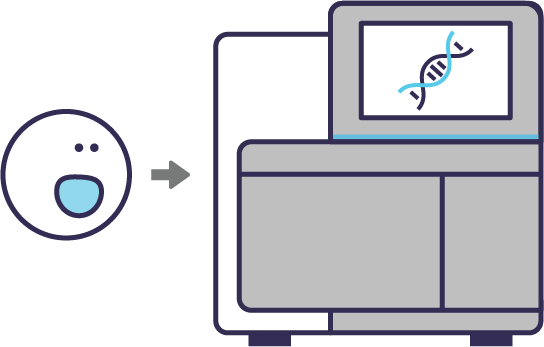
Isolated cell classes are validated with downstream tools such as multi-omics analyses and functional assays.
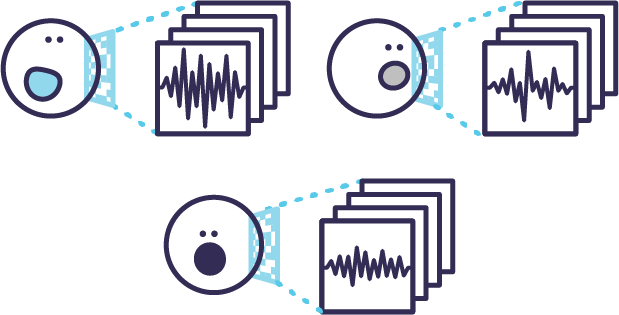
Morphological profiles are generated for each cell by Ghost Cytometry
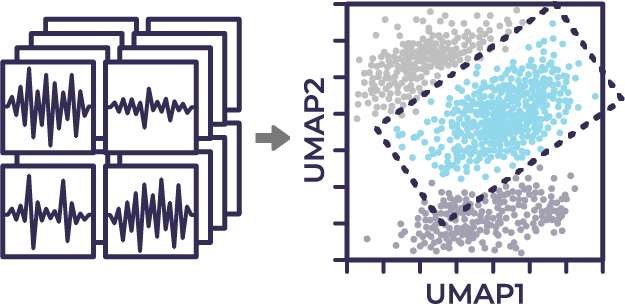
Morphological profiles are collapsed using dimensional reduction methods such as UMAP. Users create gates to identify potentially unique subpopulations.
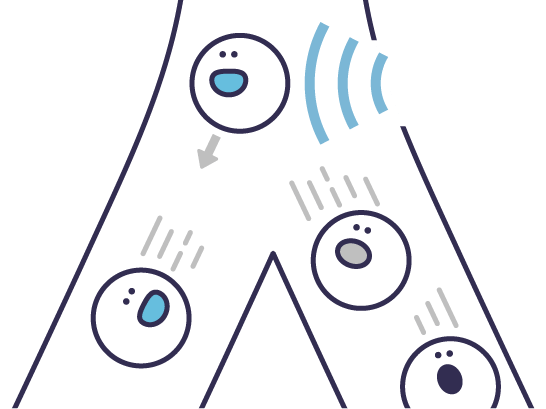
Classification models for subpopulations are created to enable real time live cell sorting.

Isolated cell classes are validated with downstream tools such as multi-omics analyses and functional assays.

1100 Island Drive STE 203
Redwood City, CA 94065
7-3-1 Hongo, Bunkyo, Tokyo
シンクサイト株式会社
東京都文京区本郷7-3-1